Protein Homeostasis and Dynamics Group
We are a molecular systems biology lab focused on understanding protein dynamics and cellular processes through proteomics and bioinformatics approaches.
Research Focus
We take an interdisciplinary approach that combines proteomics technology with biological inquiries
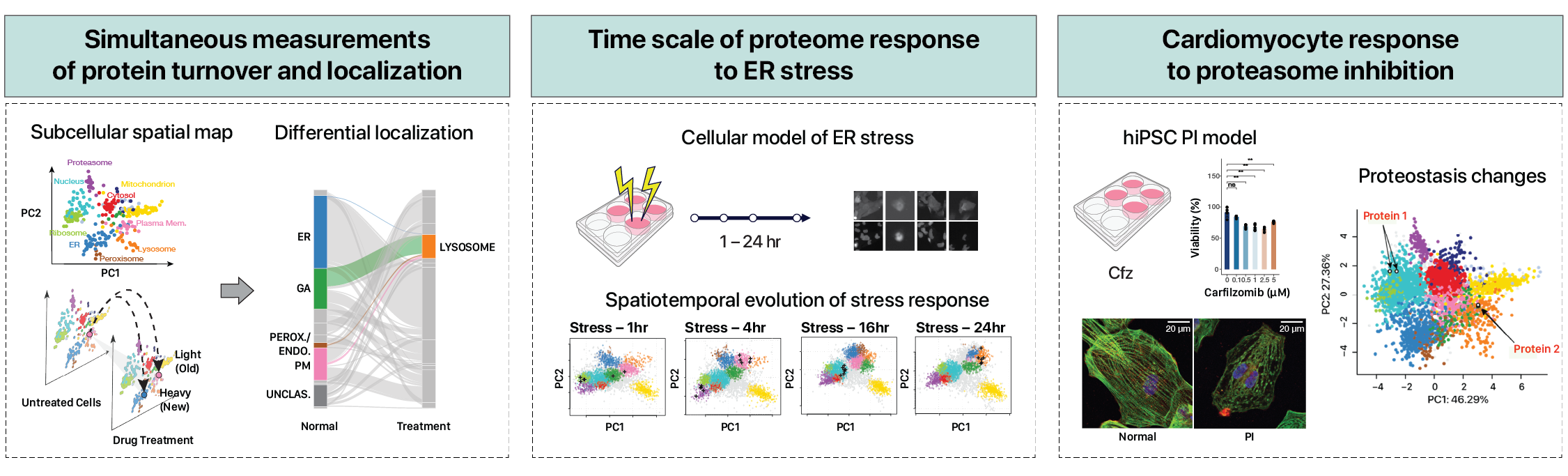
Spatiotemporal proteomics
Understanding the subcellular regulations of protein homeostasis
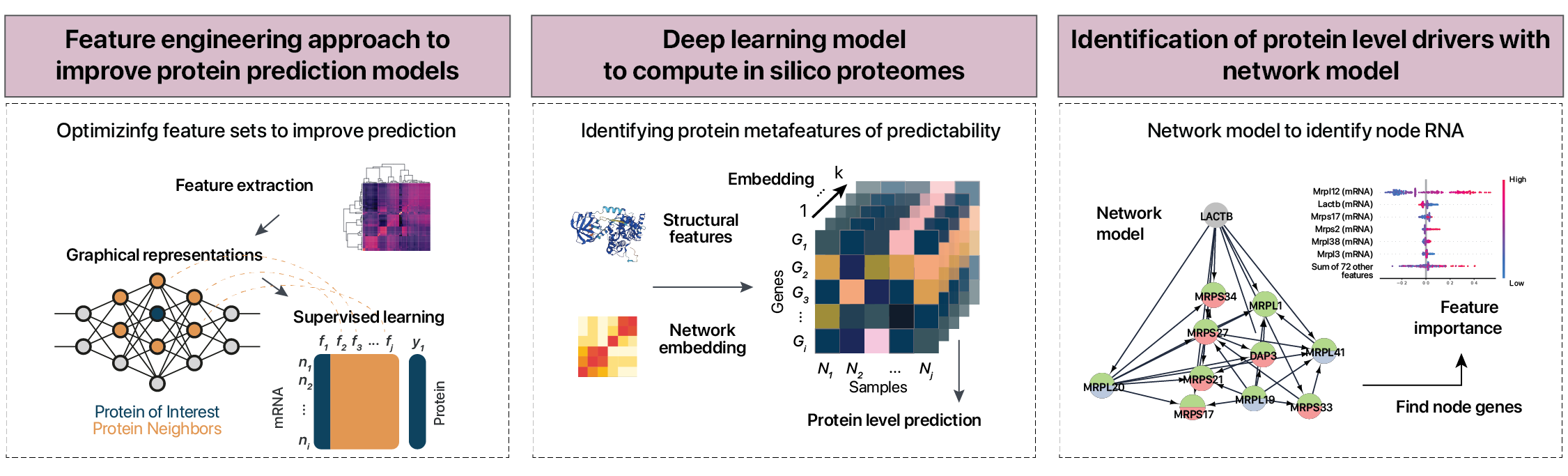
RNA protein correlations
Machine learning and deep learning methods to predict proteins from mRNA
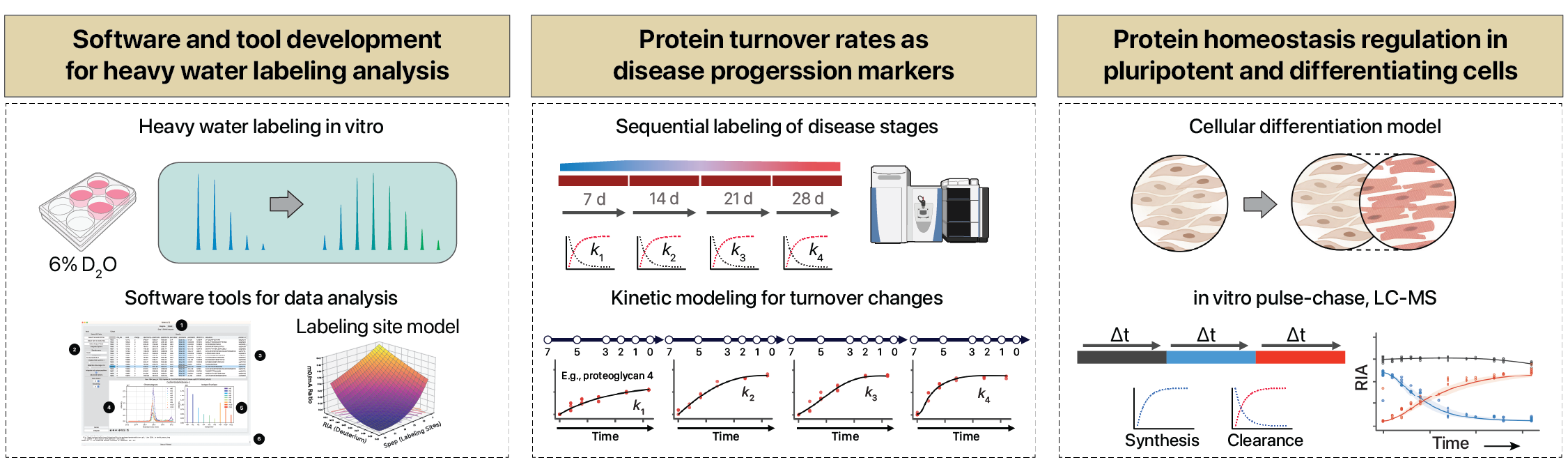
Protein turnover kinetics
We develop methods to measure protein synthesis and degradation flux

Principal Investigator
Edward Lau
Ed Lau is an Associate Professor in the Department of Medicine of the University of Colorado School of Medicine. He is also a faculty member of the Integrated Physiology, Pharmacology & Molecular Medicine, and Cell, Stem Cell & Development PhD programs on campus. Ed received his BA degree in molecular …
Research Interests: Protein turnover, kinetic models
Data & Tools
Open resources from our laboratory
Interactive Data
Lab Updates
Recent news and announcements